Path Analysis
Path analysis is a type of statistical method to investigate the direct and indirect relationship among a set of exogenous (independent, predictor, input) and endogenous (dependent, output) variables. Path analysis can be viewed as generalization of regression and mediation analysis where multiple input, mediators, and output can be used. The purpose of path analysis is to study relationships among a set of observed variables, e.g., estimate and test direct and indirect effects in a system of regression equations and estimate and test theories about the absence of relationships
Path diagrams
Path analysis is often conducted based on path diagrams. Path diagram represents a model using shapes and paths. For example, the diagram below portrays the multiple regression model $Y=\beta_0 + \beta_X X + \beta_W W + \beta_Z Z + e$.
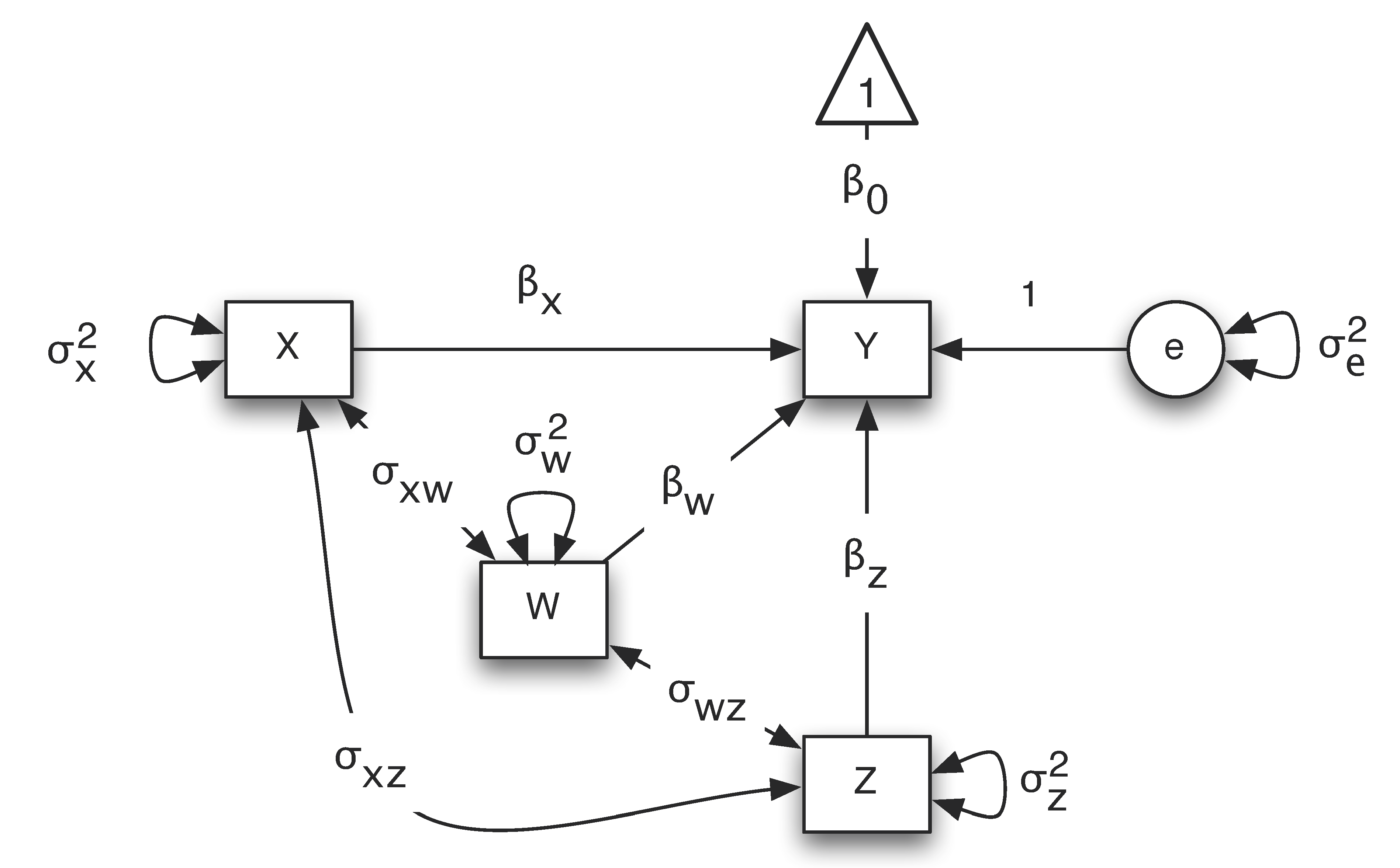
In a path diagram, different shapes and paths have different meanings:
- Squares or rectangular boxes: observed or manifest variables
- Circles or ovals: errors, factors, latent variables
- Single-headed arrows: linear relationship between two variables. Starts from an independent variable and ends on a dependent variable.
- Double-headed arrows: variance of a variable or covariance between two variables
- Triangle: a constant variable, usually a vector of ones
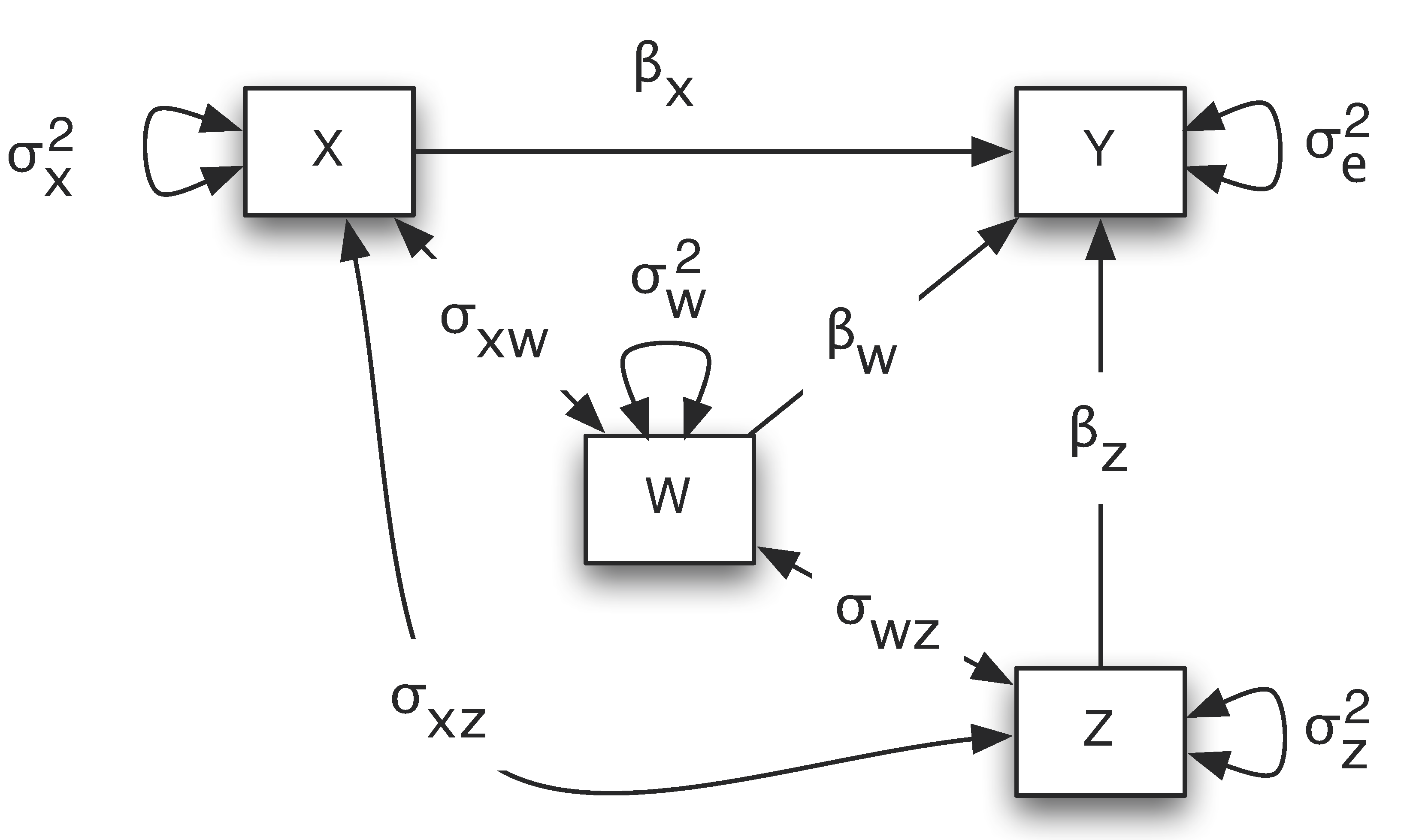
A simplified path diagram is often used in practice in which the intercept term is removed and the residual variances are directly put on the outcome variables. For example, for the regression example, the path diagram is shown below.
In python, path analysis can be conducted using the Python library semopy. semopy stands for Structural Equation Models Optimization in Python and is a powerful statistical technique that combines elements of regression, factor analysis, and path analysis to analyze relationships between observed (measured) and latent (unobserved) variables. The library is designed to be user-friendly and easy to use. It provides a simple interface to specify a model and estimate the parameters. The library also provides a variety of tools to evaluate the model fit, such as the chi-square test, RMSEA, CFI, and TLI.
We now show how to conduct path analysis using several examples.
Example 1. Mediation analysis -- Test the direct and indirect effects
The NLSY data include three variables – mother's education (ME), home environment (HE), and child's math score. Assume we want to test whether home environment is a mediator between mother’s education and child's math score. The path diagram for the mediation model is:
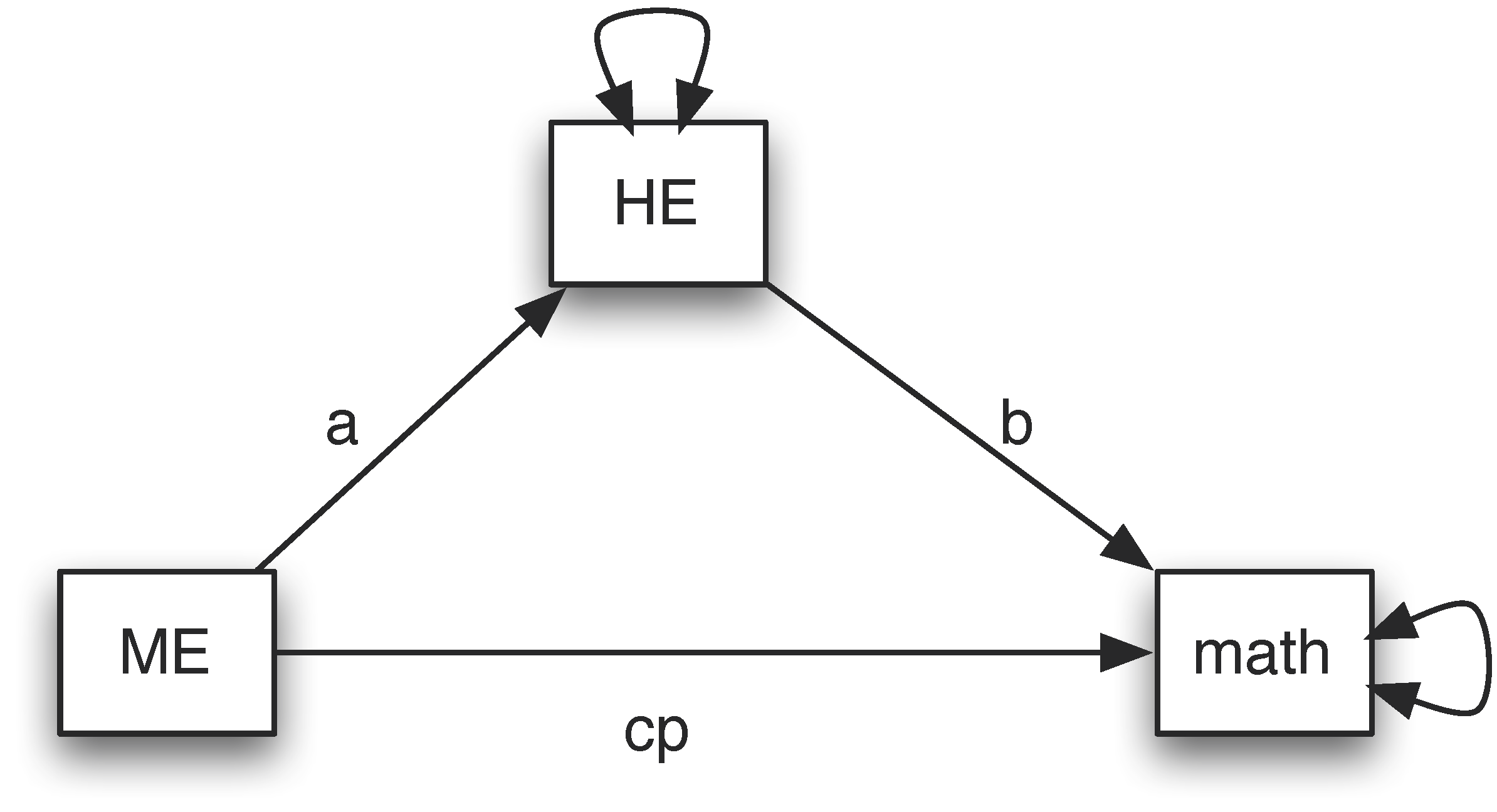
To estimate the paths in the model, we use the Python library semopy. To specify the mediation model, we follow the rules below. First, a model can be specified using strings. The triple-quoted strings, which are strings enclosed in three single quotes (''') or three double quotes (""") are used. The quotes surround a model. Second, to specify the regression relationship, we use a symbol ~. The variable on the left is the outcome and the ones on the right are predictors or covariates. Third, to specify the covariance relationship, we use a symbol ~~. Fourth, parameter names can be used for paths in model specification such as a, b and cp.
With the specified model in string, we can compile is using the Model() function. The fit() function is used to estimate the parameters. The inspect() function is used to get the parameter estimates. To get the fit statistics, the calc_stats() function is used. For example, for the mediation example, the input and output are given below.
>>> import pandas as pd
>>> import semopy as sem
>>>
>>> nlsy = pd.read_csv('https://advstats.psychstat.org/data/nlsy.csv')
>>> nlsy.head()
ME HE math
0 12 6 6
1 15 6 11
2 9 5 3
3 8 3 6
4 9 6 11
>>>
>>> # Define the path model using the function Model
>>> med = '''
... math ~ HE + ME
... HE ~ ME
... ME ~~ ME
... '''
>>>
>>> # Create the SEM model
>>> model = sem.Model(med)
>>>
>>> # Fit the model to data
>>> model.fit(nlsy)
SolverResult(fun=np.float64(8.785567562341612e-08), success=True, n_it=19, x=array([ 0.46453692, 0.46244455, 0.1391007 , 3.9951032 , 2.72321485,
20.61697808]), message='Optimization terminated successfully', name_method='SLSQP', name_obj='MLW')
>>>
>>> # Get parameter estimates and model fit statistics
>>> params = model.inspect()
>>> print(params)
lval op rval Estimate Std. Err z-value p-value
0 HE ~ ME 0.139101 0.042864 3.245184 0.001174
1 math ~ HE 0.464537 0.142851 3.251888 0.001146
2 math ~ ME 0.462445 0.119602 3.866518 0.000110
3 ME ~~ ME 3.995103 0.293330 13.619838 0.000000
4 HE ~~ HE 2.723215 0.199945 13.619838 0.000000
5 math ~~ math 20.616978 1.513746 13.619838 0.000000
>>>
>>> # Check model fit
>>> fit_stats = sem.calc_stats(model)
>>> print(fit_stats.T)
Value
DoF 0.000000e+00
DoF Baseline 3.000000e+00
chi2 3.259446e-05
chi2 p-value NaN
chi2 Baseline 3.978475e+01
CFI 9.999991e-01
GFI 9.999992e-01
AGFI NaN
NFI 9.999992e-01
TLI NaN
RMSEA inf
AIC 1.200000e+01
BIC 3.549721e+01
LogLik 8.785568e-08
>>>
>>> # mediation effect
>>> print(params.loc[0, 'Estimate'] * params.loc[1, 'Estimate'])
0.06461740868004863
>>>
>>> # plot the model
>>> # sem.semplot(med, 'mediation0.svg') ## without coefficients
>>> sem.semplot(model, 'mediation.svg') ## with coefficients
- An individual path can be tested. For example, the coefficient from ME to HE is 0.139, which is significant based on the z-test.
- The residual variance parameters are also automatically estimated.
The mediation effect can be calculated using the parameter estimates. The mediation effect is the product of the path from ME to HE and the path from HE to math. For testing the effect, we will use the bootstrap method.
Example 2. Testing a theory of no direct effect
Assume we hypothesize that there is no direct effect from ME to math. To test the hypothesis, we can fit a model illustrated below.
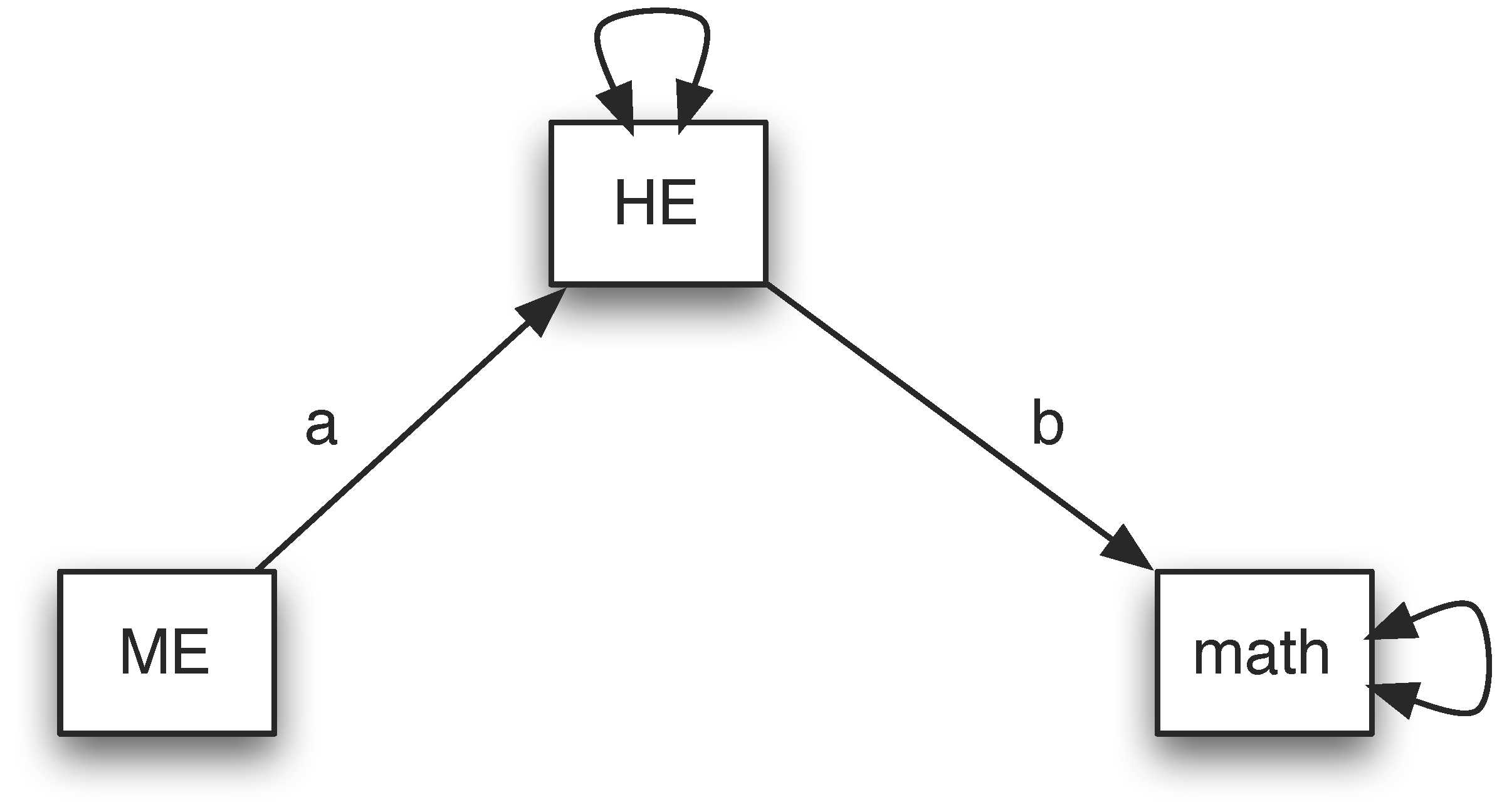
The input and output of the analysis are given below. To evaluate the hypothesis, we can check the model fit. The null hypothesis is “\(H_{0}\): The model fits the data well or the model is supported”. The alternative hypothesis is “\(H_{1}\): The model does not fit the data or the model is rejected”. The model with the direct effect fits the data perfectly. Therefore, if the current model also fits the data well, we fail to reject the null hypothesis. Otherwise, we reject it. The test of the model can be conducted based on a chi-squared test. From the output, the Chi-square is 14.676 with 1 degree of freedom. The p-value is about 0. Therefore, the null hypothesis is rejected. This indicates that the model without direct effect is not a good model.
>>> import pandas as pd
>>> import semopy as sem
>>>
>>> nlsy = pd.read_csv('https://advstats.psychstat.org/data/nlsy.csv')
>>> nlsy.head()
ME HE math
0 12 6 6
1 15 6 11
2 9 5 3
3 8 3 6
4 9 6 11
>>>
>>> # Define the path model using the function Model
>>> med = '''
... math ~ HE
... HE ~ ME
... ME ~~ ME
... '''
>>>
>>> # Create the SEM model
>>> model = sem.Model(med)
>>>
>>> # Fit the model to data
>>> model.fit(nlsy)
SolverResult(fun=np.float64(0.039558178472407945), success=True, n_it=19, x=array([ 0.55679084, 0.13915528, 3.9951032 , 2.72351785, 21.45130121]), message='Optimization terminated successfully', name_method='SLSQP', name_obj='MLW')
>>>
>>> # Get parameter estimates and model fit statistics
>>> params = model.inspect()
>>> print(params)
lval op rval Estimate Std. Err z-value p-value
0 HE ~ ME 0.139155 0.042866 3.246277 0.001169
1 math ~ HE 0.556791 0.143679 3.875248 0.000107
2 ME ~~ ME 3.995103 0.293330 13.619838 0.000000
3 HE ~~ HE 2.723518 0.199967 13.619838 0.000000
4 math ~~ math 21.451301 1.575004 13.619838 0.000000
>>>
>>> # Check model fit
>>> fit_stats = sem.calc_stats(model)
>>> print(fit_stats.T)
Value
DoF 1.000000
DoF Baseline 3.000000
chi2 14.676084
chi2 p-value 0.000128
chi2 Baseline 39.784755
CFI 0.628213
GFI 0.631113
AGFI -0.106661
NFI 0.631113
TLI -0.115360
RMSEA 0.192256
AIC 9.920884
BIC 29.501894
LogLik 0.039558
Example 3: A more complex path model
Path analysis can be used to test more complex theories. In this example, we look at how age and education influence EPT using the ACTIVE data. Both age and education may influence EPT directly or through memory and reasoning ability. Therefore, we can fit a model shown below.
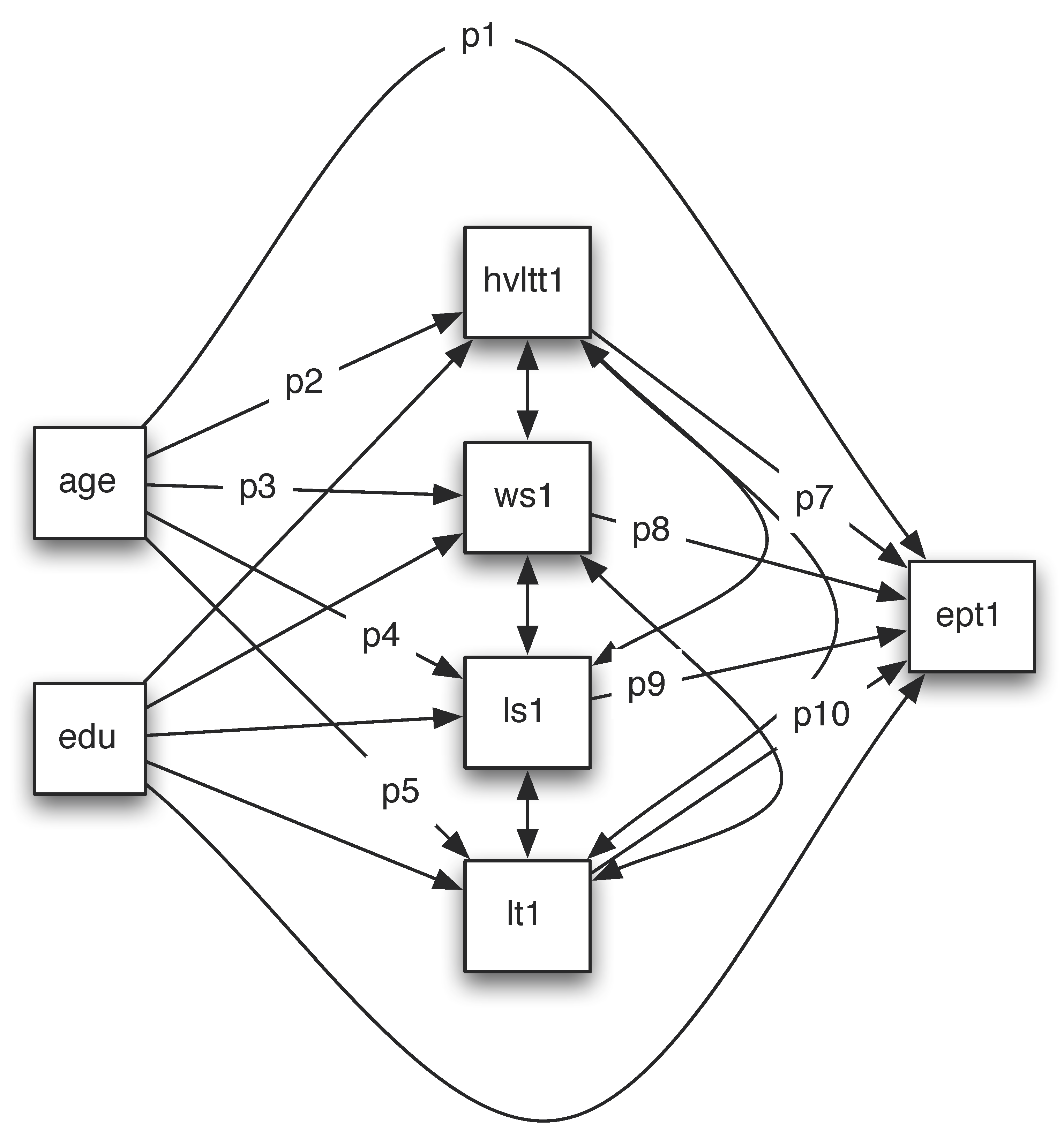
Suppose we want to test the total effect of age on EPT and its indirect effect. The direct effect is the path from age to ept1 directly, denoted by p1. One indirect path goes through hvltt1, that is p2*p7. The second indirect effect through ws1 is p3*p8. The third indirect effect through ls1 is p4*p9. The last indirect effect through lt1 is p5*p10. The total indirect effect is p2*p7+p3*p8+p4*p9+p5*p10. The total effect is the sum of them p1+p2*p7+p3*p8+p4*p9+p5*p10.
The output from such a model is given below. From it, we can see that the indirect effect ind1=p2*p7 is significant. The total indirect (indirect) from age to EPT is also significant. Finally, the total effect (total) from age to EPT is significant.
>>> import pandas as pd
>>> import semopy as sem
>>>
>>> active = pd.read_csv('https://advstats.psychstat.org/data/active.full.csv')
>>> active.head()
group training gender age edu mmse ... ept5 hvltt6 ws6 ls6 lt6 ept6
0 3 1 2 67 13 27 ... 24 23 8 12 6 21
1 4 0 2 69 12 28 ... 22 28 17 16 7 17
2 1 1 2 77 13 26 ... 21 18 10 13 6 20
3 4 0 2 79 12 30 ... 9 21 12 14 6 11
4 4 0 2 78 13 27 ... 14 19 4 7 3 16
[5 rows x 36 columns]
>>>
>>> # Define the path model using the function Model
>>> med = '''
... hvltt1 ~ age + edu
... ws1 ~ age + edu
... ls1 ~ age + edu
... lt1 ~ age + edu
... ept1 ~ age + edu + hvltt1 + ws1 + ls1 + lt1
... age ~~ age
... edu ~~ edu
... age ~~ edu
... hvltt1 ~~ ws1 + ls1 + lt1
... ws1 ~~ ls1 + lt1
... ls1 ~~ lt1
... '''
>>>
>>> # Create the SEM model
>>> model = sem.Model(med)
>>>
>>> # Fit the model to data
>>> model.fit(active, obj='MLW')
SolverResult(fun=np.float64(2.696444668259801e-05), success=True, n_it=69, x=array([-1.60949969e-01, 4.29025011e-01, -2.25689229e-01, 7.04332936e-01,
-2.76307048e-01, 8.77173999e-01, -8.54662686e-02, 3.94002008e-01,
1.37129234e-02, 4.48298951e-01, 2.02464085e-01, 1.95712061e-01,
2.45621990e-01, 1.50610194e-01, 2.64752022e+01, -9.19226170e-01,
6.75398229e+00, 7.60103819e+00, 8.48481022e+00, 2.86800334e+00,
2.06332206e+01, 1.65720608e+01, 5.70004236e+00, 1.95461570e+01,
6.56166923e+00, 2.40035114e+01, 1.21075359e+01, 6.16833077e+00]), message='Optimization terminated successfully', name_method='SLSQP', name_obj='MLW')
>>>
>>> # Get parameter estimates and model fit statistics
>>> params = model.inspect()
>>> print(params)
lval op rval Estimate Std. Err z-value p-value
0 hvltt1 ~ age -0.160950 0.026512 -6.070732 1.273284e-09
1 hvltt1 ~ edu 0.429025 0.052492 8.173218 2.220446e-16
2 ws1 ~ age -0.225689 0.025805 -8.746088 0.000000e+00
3 ws1 ~ edu 0.704333 0.051090 13.786096 0.000000e+00
4 ls1 ~ age -0.276307 0.028596 -9.662472 0.000000e+00
5 ls1 ~ edu 0.877174 0.056617 15.493244 0.000000e+00
6 lt1 ~ age -0.085466 0.014496 -5.895825 3.728135e-09
7 lt1 ~ edu 0.394002 0.028701 13.728042 0.000000e+00
8 ept1 ~ age 0.013713 0.021229 0.645953 5.183095e-01
9 ept1 ~ edu 0.448299 0.045098 9.940634 0.000000e+00
10 ept1 ~ hvltt1 0.202464 0.025110 8.063206 6.661338e-16
11 ept1 ~ ws1 0.195712 0.037700 5.191269 2.088653e-07
12 ept1 ~ ls1 0.245622 0.034545 7.110133 1.159295e-12
13 ept1 ~ lt1 0.150610 0.050866 2.960923 3.067187e-03
14 age ~~ age 26.475202 1.121790 23.600847 0.000000e+00
15 age ~~ edu -0.919226 0.401588 -2.288978 2.208062e-02
16 edu ~~ edu 6.753982 0.286175 23.600847 0.000000e+00
17 hvltt1 ~~ ws1 7.601038 0.643345 11.814879 0.000000e+00
18 hvltt1 ~~ ls1 8.484810 0.713591 11.890305 0.000000e+00
19 hvltt1 ~~ lt1 2.868003 0.348758 8.223484 2.220446e-16
20 hvltt1 ~~ hvltt1 20.633221 0.874258 23.600847 0.000000e+00
21 ws1 ~~ ls1 16.572061 0.817125 20.280947 0.000000e+00
22 ws1 ~~ lt1 5.700042 0.370668 15.377763 0.000000e+00
23 ws1 ~~ ws1 19.546157 0.828197 23.600847 0.000000e+00
24 ls1 ~~ lt1 6.561669 0.414197 15.841896 0.000000e+00
25 ls1 ~~ ls1 24.003511 1.017061 23.600847 0.000000e+00
26 lt1 ~~ lt1 6.168331 0.261361 23.600847 0.000000e+00
27 ept1 ~~ ept1 12.107536 0.513013 23.600847 0.000000e+00
>>>
>>> # Check model fit
>>> fit_stats = sem.calc_stats(model)
>>> print(fit_stats.T)
Value
DoF 0.000000
DoF Baseline 21.000000
chi2 0.030038
chi2 p-value NaN
chi2 Baseline 3349.940000
CFI 0.999991
GFI 0.999991
AGFI NaN
NFI 0.999991
TLI NaN
RMSEA inf
AIC 55.999946
BIC 196.439894
LogLik 0.000027
To cite the book, use:
Zhang, Z. & Wang, L. (2017-2026). Advanced statistics using R. Granger, IN: ISDSA Press. https://doi.org/10.35566/advstats. ISBN: 978-1-946728-01-2.
To take the full advantage of the book such as running analysis within your web browser, please subscribe.
